Motivation
[htyoutube controls=”1″ start =”10″ end=”40″ thumbnailresolution=”recommended” thumbnail=””]7Hww31vZSyA[/htyoutube]
Data scientists often work in iterative and exploratory environments. Thus, there is often a focus on rapid results rather than creating maintainable or scalable code.
However, data scientists must avoid writing poor code for the following reasons:
- Reduced code readability: Badly written code can be difficult to read and understand, making it harder for both the original author and other team members to maintain or modify the code in the future.
- Increased chances of introducing bugs: Poorly structured or inefficient code is more prone to errors, potentially affecting the accuracy of analyses or models.
- Integration challenges: Badly written code can hinder integration with production systems and handovers to other team members, including data engineers and machine learning engineers.
To write better code in data science projects, it’s crucial to recognize and address common bad practices, which may include:
- Excessive use of Jupyter Notebooks
- Vague variable names
- Redundant code
- Duplicated code segments
- Frequent use of global variables
- Lack of proper code testing

These bad practices make the code less readable, reusable, and maintainable.
To illustrate these issues, we will examine the How I made top 0.3% on a Kaggle competition notebook that participates in the House Prices — Advanced Regression Techniques competition on Kaggle.
I selected this notebook because it showcases coding practices that mirror common mistakes observed in the code of data scientists I’ve collaborated with. By examining this notebook, we can gain valuable insights into the pitfalls to avoid as data scientists.
The Excessive Use of Jupyter Notebooks
Problem
Jupyter Notebooks offer an interactive environment for code execution, visualization, and immediate feedback, making them valuable for exploratory analysis and proof of concept.
However, it is not ideal for data scientists to use Jupyter Notebooks for production-related tasks like feature engineering and model training for several reasons.
Dependency Issues in Cell Execution
Firstly, some cells may depend on the output of previous cells, and executing them in a different order can cause errors or inconsistencies in the dependent cells.
In the provided example, executing cell 16 before cell 18 results in the removal of two outliers.


Number of rows dropped: 2
However, executing cell 18 before cell 16 results in the removal of three outliers.


Number of rows dropped: 3
Performance Concerns
Secondly, notebooks frequently contain a combination of visualization and analysis code, as well as production code. This blend of code in a single notebook can lead to resource-intensive tasks that may negatively impact the performance of the production system.
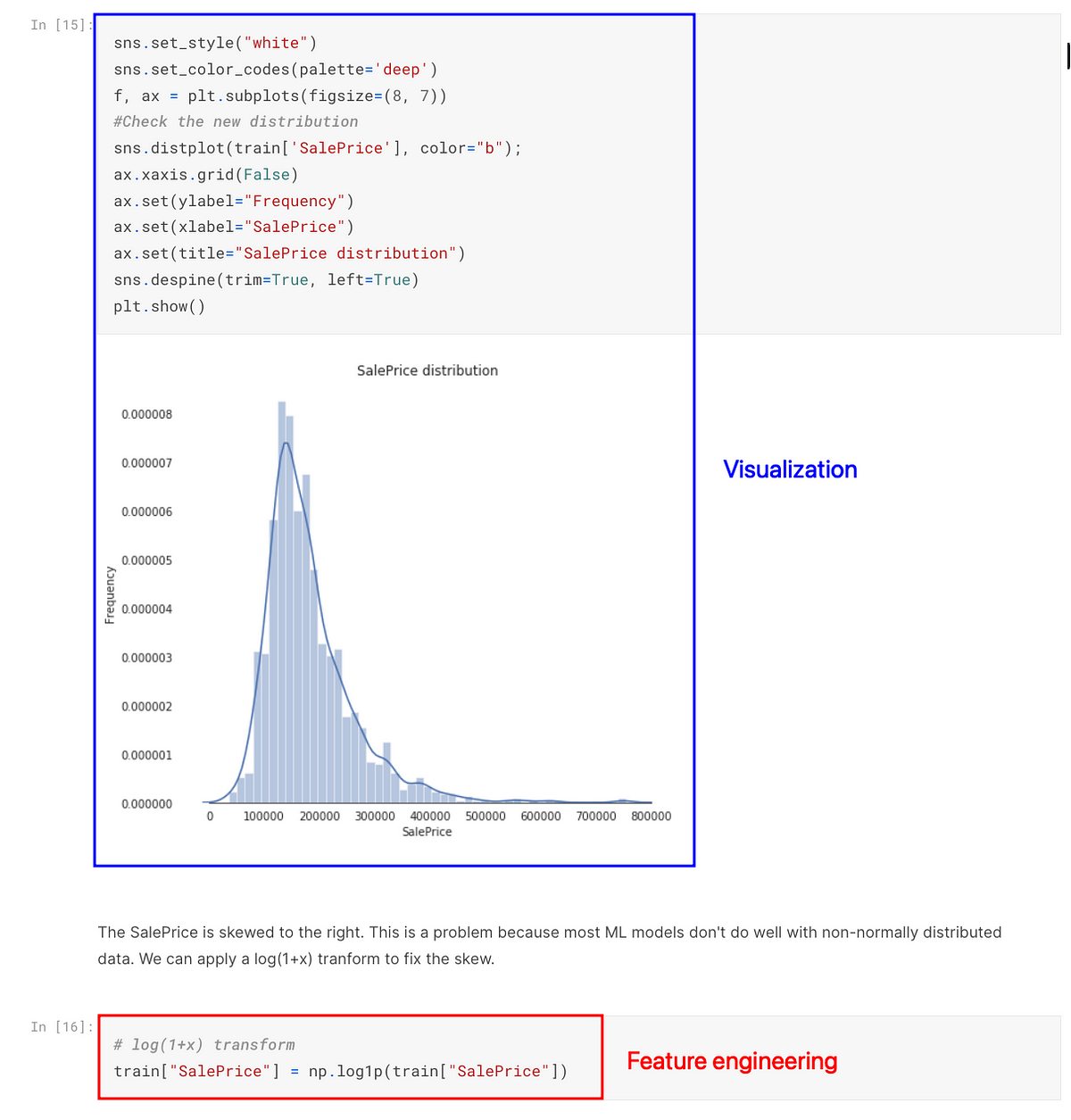
Solution
Use notebooks for EDA and analysis, while using Python scripts for feature engineering and machine learning model training.
To further organize your project, create a notebook for data analysis before feature engineering, and another notebook to analyze intermediate data after feature engineering.
.
├── data/
│ ├── raw
│ ├── intermediate
│ └── final
├── notebooks/
│ ├── pre_processing.ipynb
│ └── post_processing.ipynb
└── src/
├── __init__.py
├── process_data.py
└── train_model.pyThis approach enables the use of Python scripts in various projects while maintaining a clean and organized notebook.


Vague Variable Names
Problem
In the following code snippet, the meanings of the variables res ,ls , l , and m are unclear, making it difficult for reviewers to understand the code’s logic and potentially leading to misuse of the variables.

Solution
Use descriptive and meaningful variable names that convey the purpose and contents of the variables.
def add_log_transform_columns(data, columns):
num_columns = data.shape[1]
for column in columns:
transformed_column = pd.Series(np.log(1.01 + data[column])).values
data = data.assign(new_column=transformed_column)
data.columns.values[num_columns] = column + '_log'
num_columns += 1
return dataRedundant Code
Problem
Reduce Code Readability
Redundant code can make the code less readable.
In the notebook, the YrSold column undergoes unnecessary conversions between integer and string types.
Initially, the YrSold column is represented as an integer:

Subsequently, the code converts the YrSold column to a string:

Then, the YrSold column is temporarily transformed back to an integer:
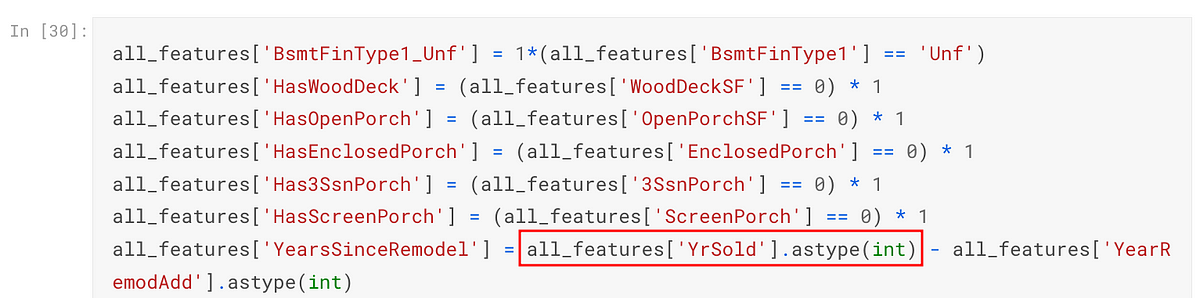
Finally, the YrSold column is converted back to an integer along with other categorical columns using one-hot encoding:

all_features.filter(regex="YrSold").dtypes
"""
YrSold_2006 int64
YrSold_2007 int64
YrSold_2008 int64
YrSold_2009 int64
YrSold_2010 int64
"""These unnecessary conversions can make it difficult for authors and maintainers to keep track of the data type of a column, which can result in the incorrect usage of the column.
Negative Performance Impact
Redundant code can also impact performance by introducing unnecessary computational overhead.
In the provided code, the author unnecessarily uses pd.DataFrame(df) twice to create two copies of a DataFrame. However, creating copies of the DataFrame is unnecessary as the objective is solely to retrieve the column names.

If the original DataFrame is large, creating new DataFrames can be computationally expensive.
Solution
Keep your code short and to the point. Remove unnecessary lines of code that don’t add value to your program.
For example, we can rewrite the code above to directly obtain the columns from the original DataFrame.
import pandas as pd
def percent_missing(df):
columns = list(df)
dict_x = {}
for i in range(0, len(columns)):
dict_x.update({columns[i]: round(df_copy[columns[i]].isnull().mean() * 100, 2)})
return missing_percentagesDuplicated Code Segments
Problem
Code duplication increases the maintenance burden.
The code 1 if x > 0 else 0 is reused multiple times. Any modifications or updates, such as changing it to 1 if x < 0 else 0, would require making the same change in every instance of the duplicated code. This process can be both time-consuming and error-prone.

Solution
Encapsulate duplicated code in functions or classes to improve code reuse and maintainability.
For example, we can create a function called is_positive that encapsulates the code snippet 1 if x > 0 else 0.
def is_positive(column):
return 1 if column > 0 else 0
all_features['haspool'] = all_features['PoolArea'].apply(is_positive)
all_features['has2ndfloor'] = all_features['2ndFlrSF'].apply(is_positive)
all_features['hasgarage'] = all_features['GarageArea'].apply(is_positive)
all_features['hasbsmt'] = all_features['TotalBsmtSF'].apply(is_positive)
all_features['hasfireplace'] = all_features['Fireplaces'].apply(is_positive)Frequent Use of Global Variables
Problem
The usage of global variables can lead to confusion and difficulties in understanding how and where the values are modified.
In the following code, X, train_labels, and kf are global variables that are defined in different parts of the codebase.




When looking at the function call, maintainers may incorrectly assume that the cv_rmse function can be invoked with only the model variable defined:
model = LinearRegression()
rmse_scores = cv_rsme(model), … but in reality, the function requires X, train_labels, and kf to be defined as well.
Traceback (most recent call last):
File "/Users/khuyentran/software-engineering-for-data-scientists/variables/global_variables/main.py", line 20, in <module>
scores = cv_rsme(model)
^^^^^^^^^^^^^^
File "/Users/khuyentran/software-engineering-for-data-scientists/variables/global_variables/main.py", line 13, in cv_rsme
return np.sqrt(-cross_val_score(model, X, train_labels, scoring='neg_mean_squared_error', cv=kf))
^
NameError: name 'X' is not definedSolution
Instead of using global variables, pass the necessary variables as arguments to the function. This will make the function more modular and easier to test.
def cv_rmse(model, X, train_labels, kf):
rmse = np.sqrt(-cross_val_score(model, X, train_labels, scoring="neg_mean_squared_error", cv=kf))
return rmse
model = LinearRegression()
rmse_scores = cv_rmse(model, X=..., train_labels=..., kf=...)
print((rmse_scores.mean(), rmse_scores.std()))
# (1.092857142857143, 0.5118992247762547)Lack of Proper Code Testing
Hidden Code Issues
Problem
Untested code can yield unexpected results, even if the output seems correct.
In the code example, using the create_booleans function on integers should turn them into 0s and 1s. The output appears correct, with 0s and 1s, but it’s actually wrong. Non-zero values should be 1, and zeros should be 0.
import pandas as pd
data = {
'WoodDeckSF': [150, 0, 80, 120, 200],
'OpenPorchSF': [30, 40, 0, 20, 60],
'EnclosedPorch': [0, 20, 10, 0, 30],
'3SsnPorch': [0, 0, 0, 15, 0],
'ScreenPorch': [0, 0, 25, 0, 40]
}
all_features = pd.DataFrame(data)
all_features['HasWoodDeck'] = (all_features['WoodDeckSF'] == 0) * 1
all_features['HasOpenPorch'] = (all_features['OpenPorchSF'] == 0) * 1
all_features['HasEnclosedPorch'] = (all_features['EnclosedPorch'] == 0) * 1
all_features['Has3SsnPorch'] = (all_features['3SsnPorch'] == 0) * 1
all_features['HasScreenPorch'] = (all_features['ScreenPorch'] == 0) * 1
all_features.iloc[:, -5:]
# The results are wrong
"""
HasWoodDeck HasOpenPorch HasEnclosedPorch Has3SsnPorch HasScreenPorch
0 0 0 1 1 1
1 1 0 0 1 1
2 0 1 0 1 0
3 0 0 1 0 1
4 0 0 0 1 0
"""Relying on inaccurate outcomes can result in faulty analyses and misleading conclusions.
Solution
With unit tests, we can specify the expected output, reducing the likelihood of overlooking bugs.
import pandas as pd
from pandas.testing import assert_series_equal
def create_booleans(feature):
return (feature == 0) * 1
def test_create_booleans():
feature = pd.Series([4, 2, 0, 1])
expected = pd.Series([1, 1, 0, 1])
actual = create_booleans(feature)
assert_series_equal(expected, actual)============================ FAILURES ============================
______________________ test_create_booleans ______________________
def test_create_booleans():
feature = pd.Series([4, 2, 0, 1])
expected = pd.Series([1, 1, 0, 1])
actual = create_booleans(feature)
> assert_series_equal(expected, actual)
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
E AssertionError: Series are different
E
E Series values are different (100.0 %)
E [index]: [0, 1, 2, 3]
E [left]: [1, 1, 0, 1]
E [right]: [0, 0, 1, 0]
E At positional index 0, first diff: 1 != 0Overlooked Edge Cases
Problem
Code may perform well in specific conditions but exhibit unexpected behaviors in others.
In this example, the code fills missing values in the MSZoning column based on the mode of values for each group in the MSSubClass column. It works as expected when MSSubClass has no NaN values.
features = pd.DataFrame(
{
"MSZoning": [1, np.nan, 2, 3, 4, 5, 6, np.nan],
"MSSubClass": ["a", "a", "a", "a", "b", "b", "b", "b"],
}
)
features['MSZoning'] = features.groupby('MSSubClass')['MSZoning'].transform(lambda x: x.fillna(x.mode()[0]))
list(features["MSZoning"])
# [1.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 4.0]However, when MSSubClass contains missing values, the code doesn’t behave as expected, leaving some missing values in MSZoning.
features = pd.DataFrame(
{
"MSZoning": [1, np.nan, 2, 3, 4, 5, 6, np.nan],
"MSSubClass": ["a", "a", np.nan, "a", np.nan, "b", "b", "b"],
}
)
features['MSZoning'] = features.groupby('MSSubClass')['MSZoning'].transform(lambda x: x.fillna(x.mode()[0]))
list(features["MSZoning"])
# [1.0, 1.0, nan, 3.0, nan, 5.0, 6.0, 5.0]Neglecting to address edge cases can lead to problems in real-world applications.
Solution
We can use unit tests to test for edge cases:
def fill_missing_values_with_mode_in_a_group(
df: pd.DataFrame, group_column: str, target_column: str
) -> pd.DataFrame:
df[target_column] = df.groupby(group_column)[target_column].transform(
lambda x: x.fillna(x.mode()[0])
)
return df
def test_fill_missing_values_with_mode_in_a_group():
data = pd.DataFrame(
{
"col1": [1, np.nan, 2, 3, 4, 5, 6, np.nan],
"col2": ["a", "a", np.nan, "a", np.nan, "b", "b", "b"],
}
)
imputed_data = fill_missing_values_with_mode_in_a_group(
df=data,
group_column="col2",
target_column="col1",
)
assert imputed_data['col1'].isnull().sum() == 0, "There are missing values in the column."AssertionError: There are missing values in the column.
assert 2 == 0AssertionError: There are missing values in the column.
assert 2 == 0… and adjust the code to account for edge cases:
def fill_missing_values_with_mode_in_a_group(
df: pd.DataFrame, group_column: str, target_column: str
) -> pd.DataFrame:
if df[group_column].isna().any():
raise ValueError(
f"The {group_column} used for grouping cannot contain null values"
)
df[target_column] = df.groupby(group_column)[target_column].transform(
lambda x: x.fillna(x.mode()[0])
)
return df
def test_fill_missing_values_with_mode_in_a_group():
with pytest.raises(ValueError):
data = pd.DataFrame(
{
"col1": [1, np.nan, 2, 3, 4, 5, 6, np.nan],
"col2": ["a", "a", np.nan, "a", np.nan, "b", "b", "b"],
}
)
imputed_data = fill_missing_values_with_mode_in_a_group(
df=data,
group_column="col2",
target_column="col1",
)Conclusion
This article discusses common challenges encountered in data science projects and provides some practical solutions to address them. Please note that this article does not exhaust all possible solutions, but rather offers a selection of strategies that may help overcome these issues.