Table of Contents
- Introduction
- What is Coiled?
- Setup
- Serverless Functions: Process Data with Any Framework
- When You Need More: Distributed Clusters with Dask
- Cost Optimization
- Environment Synchronization
- Conclusion
Introduction
A common challenge data scientists face is that local machines simply can’t handle large-scale datasets. Once your analysis reaches the 50GB+ range, you’re pushed into difficult choices:
- Sample your data and hope patterns hold
- Buy more RAM or rent expensive cloud VMs
- Learn Kubernetes and spend days configuring clusters
- Build Docker images and manage container registries
Each option adds complexity, cost, or compromises your analysis quality.
Coiled eliminates these tradeoffs. It provisions emphemeral compute clusters on AWS, GCP, or Azure using simple Python APIs. You get distributed computing power without DevOps expertise, automatic environment synchronization without Docker, and 70% cost savings through smart spot instance management.
In this article, you’ll learn how to scale Python data workflows to the cloud with Coiled:
- Serverless functions: Run pandas, Polars, or DuckDB code on cloud VMs with a simple decorator
- Parallel processing: Process multiple files simultaneously across cloud machines
- Distributed clusters: Aggregate data across files using managed Dask clusters
- Environment sync: Replicate your local packages to the cloud without Docker
- Cost optimization: Reduce cloud spending with spot instances and auto-scaling
💻 Get the Code: The complete source code and Jupyter notebook for this tutorial are available on GitHub. Clone it to follow along!
Stay Current with CodeCut
Actionable Python tips, curated for busy data pros. Skim in under 2 minutes, three times a week.
What is Coiled?
Coiled is a lightweight cloud platform that runs Python code on powerful cloud infrastructure without requiring Docker or Kubernetes knowledge. It supports four main capabilities:
- Batch Jobs: Submit and run Python scripts asynchronously on cloud infrastructure
- Serverless Functions: Execute Python functions (Pandas, Polars, PyTorch) on cloud VMs with decorators
- Dask Clusters: Provision multi-worker clusters for distributed computing
- Jupyter Notebooks: Launch interactive Jupyter servers directly on cluster schedulers
Key features across both:
- Framework-Agnostic: Works with Pandas, Polars, Dask, or any Python library
- Automatic Package Sync: Local packages replicate to cloud workers without Docker
- Cost Optimization: Spot instances, adaptive scaling, and auto-shutdown reduce spending
- Simple APIs: Decorate functions or create clusters with 2-3 lines of code
To install Coiled and Dask, run:
pip install coiled dask[complete]
Setup
First, create a free Coiled account by running this command in your terminal:
coiled login
This creates a free Coiled Hosted account with 200 CPU-hours per month. Your code runs on Coiled’s cloud infrastructure with no AWS/GCP/Azure account needed.
Note: For production workloads with your own cloud account, run coiled setup PROVIDER (setup guide).
Serverless Functions: Process Data with Any Framework
The simplest way to scale Python code to the cloud is with serverless functions. Decorate any function with @coiled.function, and Coiled handles provisioning cloud VMs, installing packages, and executing your code.
Scale Beyond Laptop Memory with Cloud VMs
Imagine you need to process a NYCTaxi dataset with 12GB compressed files (50GB+ expanded) on a laptop with only 16GB of RAM. Your machine simply doesn’t have enough memory to handle this workload.
With Coiled, you can run the exact same code on a cloud VM with 64GB of RAM by simply adding the @coiled.function decorator.
import coiled
import pandas as pd
@coiled.function(
memory="64 GiB",
region="us-east-1"
)
def process_month_with_pandas(month):
# Read 12GB file directly into pandas
df = pd.read_parquet(
f"https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2024-{month:02d}.parquet"
)
# Compute tipping patterns by hour
df["hour"] = df["tpep_pickup_datetime"].dt.hour
result = df.groupby("hour")["tip_amount"].mean()
return result
# Run on cloud VM with 64GB RAM
january_tips = process_month_with_pandas(1)
print(january_tips)
Output:
hour
0 3.326543
1 2.933899
2 2.768246
3 2.816333
4 3.132973
...
Name: tip_amount, dtype: float64
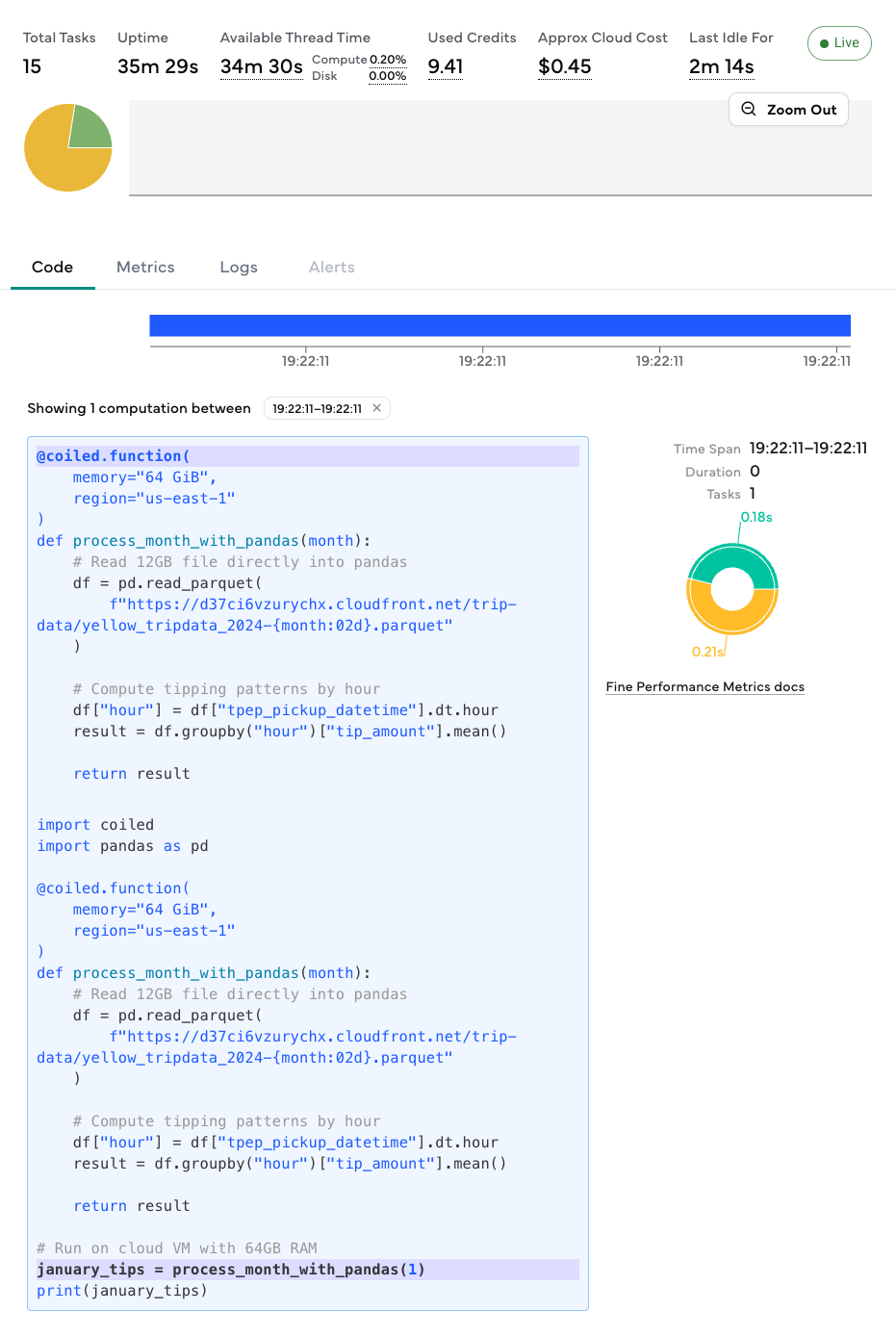
This function runs on a cloud VM with 64GB RAM, processes the entire month in memory, and returns just the aggregated result to your laptop.
You can view the function’s execution progress and resource usage in the Coiled dashboard.
Parallel Processing with .map()
By default Coiled Functions will run sequentially, just like normal Python functions. However, they can also easily run in parallel by using the .map() method.
Process all 12 months in parallel using .map():
import coiled
import pandas as pd
@coiled.function(memory="64 GiB", region="us-east-1")
def process_month(month):
df = pd.read_parquet(
f"https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2024-{month:02d}.parquet"
)
return df["tip_amount"].mean()
# Process 12 months in parallel on 12 cloud VMs
months = range(1, 13)
monthly_tips = list(process_month.map(months))
print("Average tips by month:", monthly_tips)
Output:
Average tips by month: [2.65, 2.58, 2.72, 2.68, 2.75, 2.81, 2.79, 2.73, 2.69, 2.71, 2.66, 2.62]
When you call .map() with 12 months, Coiled spins up 12 cloud VMs simultaneously, runs process_month() on each VM with a different month, then returns all results.
The execution flow:
VM 1: yellow_tripdata_2024-01.parquet → compute mean → 2.65
VM 2: yellow_tripdata_2024-02.parquet → compute mean → 2.58
VM 3: yellow_tripdata_2024-03.parquet → compute mean → 2.72
... (all running in parallel)
VM 12: yellow_tripdata_2024-12.parquet → compute mean → 2.62
↓
Coiled collects: [2.65, 2.58, 2.72, ..., 2.62]
Each VM works in complete isolation with no data sharing or coordination between them.
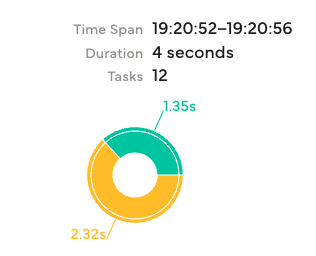
The dashboard confirms 12 tasks were executed, matching the 12 months we passed to .map().
Framework-Agnostic: Use Any Python Library
Coiled Functions aren’t limited to pandas. You can use any Python library (Polars, DuckDB, PyTorch, scikit-learn) without any additional configuration. The automatic package synchronization works for all dependencies.
Example with Polars:
Polars is a fast DataFrame library optimized for performance. It works seamlessly with Coiled:
import coiled
import polars as pl
@coiled.function(memory="64 GiB", region="us-east-1")
def process_with_polars(month):
df = pl.read_parquet(
f"https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2024-{month:02d}.parquet"
)
return (
df
.filter(pl.col("tip_amount") > 0)
.group_by("PULocationID")
.agg(pl.col("tip_amount").mean())
.sort("tip_amount", descending=True)
.head(5)
)
result = process_with_polars(1)
print(result)
Output:
shape: (5, 2)
┌──────────────┬────────────┐
│ PULocationID ┆ tip_amount │
│ --- ┆ --- │
│ i64 ┆ f64 │
╞══════════════╪════════════╡
│ 138 ┆ 4.52 │
│ 230 ┆ 4.23 │
│ 161 ┆ 4.15 │
│ 234 ┆ 3.98 │
│ 162 ┆ 3.87 │
└──────────────┴────────────┘
Example with DuckDB:
DuckDB provides fast SQL analytics directly on Parquet files:
import coiled
import duckdb
@coiled.function(memory="64 GiB", region="us-east-1")
def query_with_duckdb(month):
con = duckdb.connect()
result = con.execute(f"""
SELECT
DATE_TRUNC('hour', tpep_pickup_datetime) as pickup_hour,
AVG(tip_amount) as avg_tip,
COUNT(*) as trip_count
FROM 'https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2024-{month:02d}.parquet'
WHERE tip_amount > 0
GROUP BY pickup_hour
ORDER BY avg_tip DESC
LIMIT 5
""").fetchdf()
return result
result = query_with_duckdb(1)
print(result)
Output:
pickup_hour avg_tip trip_count
0 2024-01-15 14:00:00 4.23 15234
1 2024-01-20 18:00:00 4.15 18456
2 2024-01-08 12:00:00 3.98 12789
3 2024-01-25 16:00:00 3.87 14567
4 2024-01-12 20:00:00 3.76 16234
Coiled automatically detects your local Polars and DuckDB installations and replicates them to cloud VMs. No manual configuration needed.
When You Need More: Distributed Clusters with Dask
Serverless functions work great for independent file processing. However, when you need to combine and aggregate data across all your files into a single result, you need a Dask cluster.
For example, suppose you want to calculate total revenue by pickup location across all 12 months of data. With Coiled Functions, each VM processes one month independently:
@coiled.function(memory="64 GiB", region="us-east-1")
def get_monthly_revenue_by_location(month):
df = pd.read_parquet(
f"https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2024-{month:02d}.parquet"
)
return df.groupby("PULocationID")["total_amount"].sum()
# This returns 12 separate DataFrames, one per month
results = list(get_monthly_revenue_by_location.map(range(1, 13)))
print(f'Number of DataFrames: {len(results)}')
Output:
Number of DataFrames: 12
The problem is that you get 12 separate DataFrames that you need to manually combine.
Here’s what happens: VM 1 processes January and returns a DataFrame like:
PULocationID total_amount
138 15000
230 22000
VM 2 processes February and returns:
PULocationID total_amount
138 18000
230 19000
Each VM works independently and has no knowledge of the other months’ data. To get yearly totals per location, you’d need to write code to merge these 12 DataFrames and sum the revenue for each location.
With a Dask cluster, workers coordinate to give you one global result:
import coiled
import dask.dataframe as dd
# For production workloads, you can scale to 50+ workers
cluster = coiled.Cluster(n_workers=3, region="us-east-1")
# Read all 12 months of 2024 data
files = [
f"https://d37ci6vzurychx.cloudfront.net/trip-data/yellow_tripdata_2024-{month:02d}.parquet"
for month in range(1, 13)
]
df = dd.read_parquet(files) # Lazy: builds a plan, doesn't load data yet
# This returns ONE DataFrame with total revenue per location across all months
total_revenue = (
df.groupby("PULocationID")["total_amount"].sum().compute()
) # Executes the plan
total_revenue.head()
Output:
PULocationID
1 563645.70
2 3585.80
3 67261.41
4 1687265.08
5 602.98
Name: total_amount, dtype: float64
You can see that we got a single DataFrame with the total revenue per location across all months.
Here is what happens under the hood:
When you call .compute(), Dask executes the plan in four steps:
Step 1: Data Distribution
├─ Worker 1: [Jan partitions 1-3, Apr partitions 1-2, Jul partitions 1-3]
├─ Worker 2: [Feb partitions 1-4, May partitions 1-3, Aug partitions 1-2]
└─ Worker 3: [Mar partitions 1-3, Jun partitions 1-2, Sep-Dec partitions]
Step 2: Local Aggregation (each worker groups its data)
├─ Worker 1: {location_138: $45,000, location_230: $63,000}
├─ Worker 2: {location_138: $38,000, location_230: $55,000}
└─ Worker 3: {location_138: $50,000, location_230: $62,000}
Step 3: Shuffle (redistribute so each location lives on one worker)
├─ Worker 1: All location_230 data → $63,000 + $55,000 + $62,000
├─ Worker 2: All location_138 data → $45,000 + $38,000 + $50,000
└─ Worker 3: All other locations...
Step 4: Final Result
location_138: $133,000 (yearly total)
location_230: $180,000 (yearly total)
This shuffle-and-combine process is what makes Dask different from Coiled Functions. Workers actively coordinate and share data to produce one unified result.
Cost Optimization
Cloud costs can spiral quickly. Coiled provides three mechanisms to reduce spending:
1. Spot Instances
You can reduce cloud costs by 60-90% using spot instances. These are discounted servers that cloud providers can reclaim when demand increases. When an interruption occurs, Coiled:
- Gracefully shuts down the affected worker
- Redistributes its work to healthy workers
- Automatically launches a replacement worker
cluster = coiled.Cluster(
n_workers=50,
spot_policy="spot_with_fallback", # Use spot instances with on-demand backup
region="us-east-1"
)
Cost comparison for m5.xlarge instances:
- On-demand: $0.192/hour
- Spot: $0.05/hour
- Savings: 74%
For a 100-worker cluster:
- On-demand: $19.20/hour = $460/day
- Spot: $5.00/hour = $120/day
2. Adaptive Scaling
Adaptive scaling automatically adds workers when you have more work and removes them when idle, so you only pay for what you need. Coiled enables this with the adapt() method:
cluster = coiled.Cluster(region="us-east-1")
cluster.adapt(minimum=10, maximum=50) # Scale between 10-50 workers
Serverless functions also support auto-scaling by specifying a worker range:
@coiled.function(n_workers=[10, 300])
def process_data(files):
return results
This saves money during light workloads while delivering performance during heavy computation. No manual monitoring required.
3. Automatic Shutdown
To prevent paying for unused resources, Coiled automatically shuts down clusters after 20 minutes of inactivity by default. You can customize this with the idle_timeout parameter:
cluster = coiled.Cluster(
n_workers=20,
region="us-east-1",
idle_timeout="1 hour" # Keep cluster alive for longer workloads
)
This prevents the common mistake of leaving clusters running overnight.
Environment Synchronization
The “Works on My Machine” Problem When Scaling to Cloud
Imagine this scenario: your pandas code works locally but fails on a cloud VM because the environment has different package versions or missing dependencies.
Docker solves this by packaging your environment into a container that runs identically on your laptop and cloud VMs. However, getting it running on cloud infrastructure involves a complex workflow:
- Write a Dockerfile listing all dependencies and versions
- Build the Docker image (wait 5-10 minutes)
- Push to cloud container registry (AWS ECR, Google Container Registry)
- Configure cloud VMs (EC2/GCE instances with proper networking and security)
- Pull and run the image on cloud machines (3-5 minutes per VM)
- Rebuild and redeploy every time you add a package (repeat steps 2-5)
This Docker + cloud workflow slows down development and requires expertise in both containerization and cloud infrastructure management.
Coiled’s Solution: Automatic Package Synchronization
Coiled eliminates Docker entirely through automatic package synchronization. Your local environment replicates to cloud workers with no Dockerfile required.
Instead of managing Docker images and cloud infrastructure, you simply add a decorator to your function:
import coiled
import pandas as pd
@coiled.function(memory="64 GiB", region="us-east-1")
def process_data():
df = pd.read_parquet("s3://my-bucket/data.parquet")
# Your analysis code here
return df.describe()
result = process_data() # Runs on cloud VM with your exact package versions
What Coiled does automatically:
- Scans your local environment (pip, conda packages with exact versions)
- Creates a dependency manifest (a list of all packages and their versions)
- Installs packages on cloud workers with matching versions
- Reuses built environments when your dependencies haven’t changed
This is faster than Docker builds in most cases thanks to intelligent caching, and requires zero configuration.
Conclusion
Coiled transforms cloud computing from a multi-day DevOps project into simple Python operations. Whether you’re processing a single large file with Pandas, querying multiple files with Polars, or running distributed aggregations with Dask clusters, Coiled provides the right scaling approach for your needs.
I recommend starting simple with serverless functions for single-file processing, then scale to Dask clusters when you need truly distributed computing. Coiled removes the infrastructure burden from data science workflows, letting you focus on analysis instead of operations.
Related Tutorials
- Polars vs Pandas: Performance Benchmarks and When to Switch – Choose the right DataFrame library for your workloads
- DuckDB: Fast SQL Analytics on Parquet Files – Master SQL techniques for processing data with DuckDB
- PySpark Pandas API: Familiar Syntax at Scale – Explore Spark as an alternative to Dask for distributed processing
📚 Want to go deeper? Learning new techniques is the easy part. Knowing how to structure, test, and deploy them is what separates side projects from real work. My book shows you how to build data science projects that actually make it to production. Get the book →
Stay Current with CodeCut
Actionable Python tips, curated for busy data pros. Skim in under 2 minutes, three times a week.