Table of Contents
- Motivation
- What Makes This Article Different?
- Why Marimo?
- Getting Started
- Auto-Update Dependent Cells
- Prevent Variable Redefinition
- Enable Clean Version Control
- Build Parameterized Interactive Workflows
- Isolate Execution with Sandbox Mode
- Add Lightweight Unit Testing
- Export in Multiple Reusable Formats
- Example Workflow: From Local Editing to Online Sharing
- Final Thoughts
Motivation
Have you ever struggled to create notebooks that are both interactive and reproducible? As a data scientist, it is common to rely on interactive workflows for exploration, but this often comes at the cost of consistency and repeatability.
In traditional environments like Jupyter, running cells out of order, depending on hidden state, and modifying variables mid-stream can easily lead to confusion and bugs.
Marimo is a modern Python notebook that enforces reproducibility by design, supports interactive apps, and integrates seamlessly into your data science workflows. With Marimo, you no longer have to choose between interactivity and reproducibility. You get both in a single, consistent environment.
The source code of this article can be found here:
Stay Current with CodeCut
Actionable Python tips, curated for busy data pros. Skim in under 2 minutes, three times a week.
Key Takeaways
Here’s what you’ll learn:
- Eliminate notebook state issues with automatic dependency tracking and execution
- Version control notebooks as clean Python files instead of messy JSON
- Create interactive dashboards with built-in UI components and real-time updates
- Deploy reproducible workflows with sandbox mode and dependency isolation
- Export notebooks to multiple formats including HTML, WASM, and production scripts
Why Marimo?
Marimo solves three major issues common in traditional notebooks:
- Hidden state and out-of-order execution
Marimo enforces top-to-bottom execution with clear cell dependencies.
2. Version control and diffing
Marimo notebooks are just Python scripts, which are easy to diff, review, and manage in Git.
3. Reusability and sharing
You can export a Marimo notebook as a web app or module, making your analysis immediately interactive and reusable.
📚 For comprehensive production notebook strategies including marimo, check out Production-Ready Data Science.
Getting Started
Install Marimo using pip:
pip install marimo
Create a new notebook:
marimo new my_notebook.py
Start the notebook:
marimo edit my_notebook.py
Your notebook opens in a browser, but stays a clean .py file under the hood.
Auto-Update Dependent Cells
Problem in Traditional Notebooks
When you change an input value or upstream logic, dependent cells will not automatically update, leading to inconsistent results unless you manually re-execute them.
For example, in a Jupyter Notebook, you might have:

If you change the threshold to 30 in Cell 1 but forget to rerun Cell 2, the printed output will still reflect the old threshold:

This discrepancy can be subtle and lead to incorrect conclusions if you don’t carefully re-execute every dependent cell.
How Marimo Solves This
Another major strength of Marimo is automatic dependency tracking. When you change the output of a cell, all downstream cells that depend on its result are automatically re-executed.
This ensures your notebook is always in a consistent state, without requiring manual reruns of cells. For example:

Now if you change threshold from 50 to 30, the second cell will rerun automatically, updating the filtered output:

Marimo detects the change in threshold and ensures all dependent cells update in sync, avoiding stale results.
This live dependency resolution eliminates the need to track what to re-execute, making your workflow more robust.
Prevent Variable Redefinition
Problem in Traditional Notebooks
Variables can be silently redefined in any cell, which can cause logical errors or unintentional overwrites that are hard to trace.
For example, in a Jupyter Notebook:
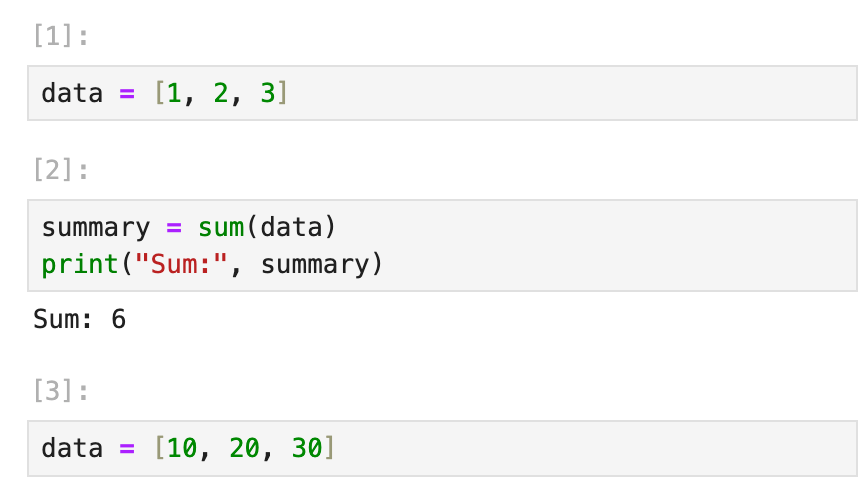
If you redefine data in Cell 3 but forget that Cell 2 depends on it, re-running Cell 2 after Cell 3 will produce a different result than originally intended, without any warning.
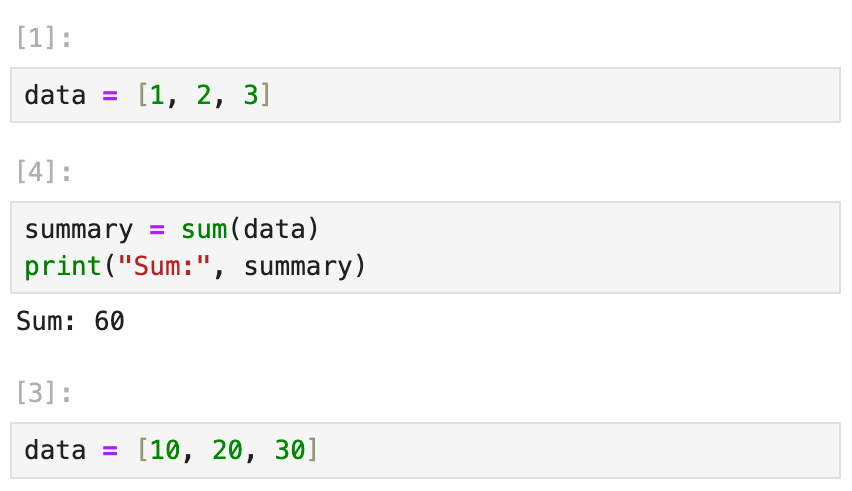
How Marimo Solves This
Marimo prevents you from accidentally redefining a variable across different cells. This eliminates bugs caused by naming collisions or silent overwrites in long notebooks.
For example, if you try to redefine data in separate cells, Marimo will raise an error, warning you that data has already been defined.
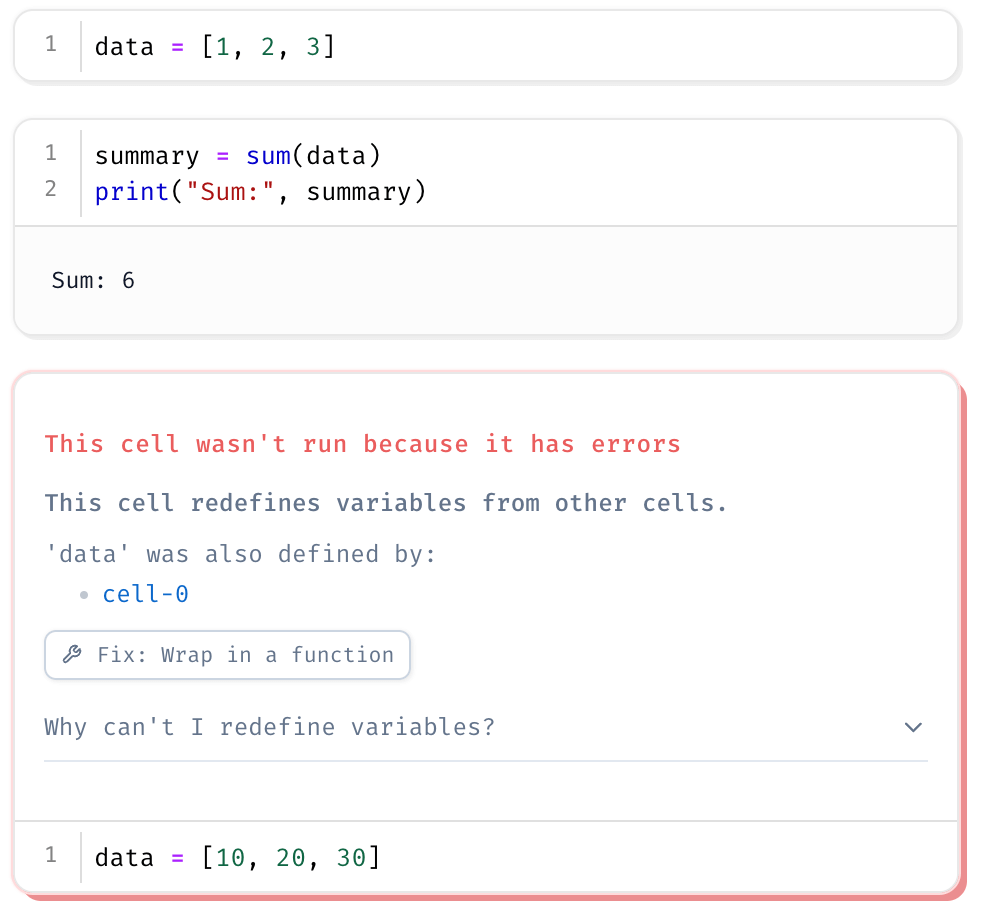
To update a variable, you must explicitly change its original definition or rename it.
This enforces clarity, making the notebook’s logic more predictable and transparent.
Enable Clean Version Control
Problem in Traditional Notebooks
.ipynb files store output alongside code in a JSON format, making diffs unreadable and code reviews difficult.
How Marimo Solves This
Since Marimo notebooks are plain Python files, they can be versioned just like any other source code.
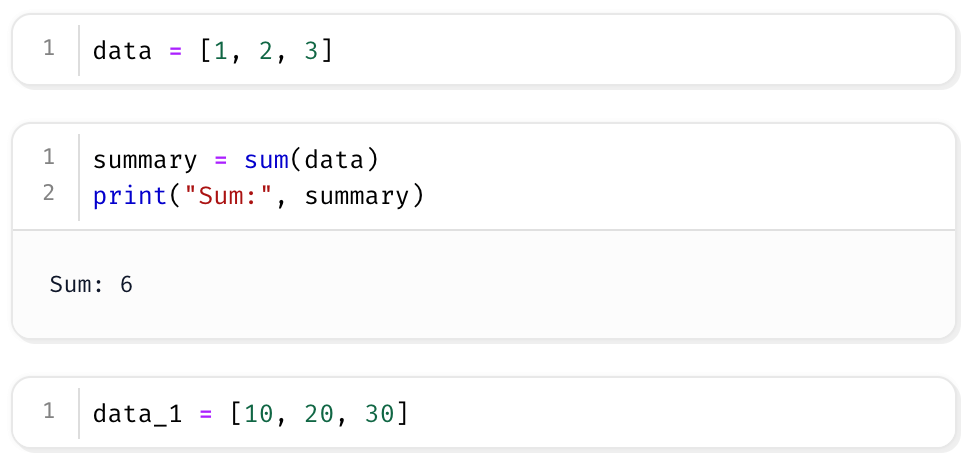
For example, this notebook:

is actually a .py file under the hood:
import marimo
__generated_with = "0.13.0"
app = marimo.App()
@app.cell
def _():
data = [1, 2, 3]
return (data,)
@app.cell
def _(data):
summary = sum(data)
print("Sum:", summary)
return
@app.cell
def _():
data_1 = [10, 20, 30]
return
@app.cell
def _():
return
if __name__ == "__main__":
app.run()
You can use Git to track changes, compare diffs, and conduct code reviews line-by-line.
Build Parameterized Interactive Workflows
Problem in Traditional Notebooks
Traditional notebook widgets (like those from ipywidgets) require boilerplate setup, often break with kernel restarts, and don’t automatically propagate changes across dependent cells, leading to brittle and inconsistent results.
How Marimo Solves This
Marimo treats interactive UI elements as native components of the notebook. They define explicit, trackable inputs that directly link to computation, ensuring that outputs are consistently and reproducibly tied to user-defined parameters.
Example:
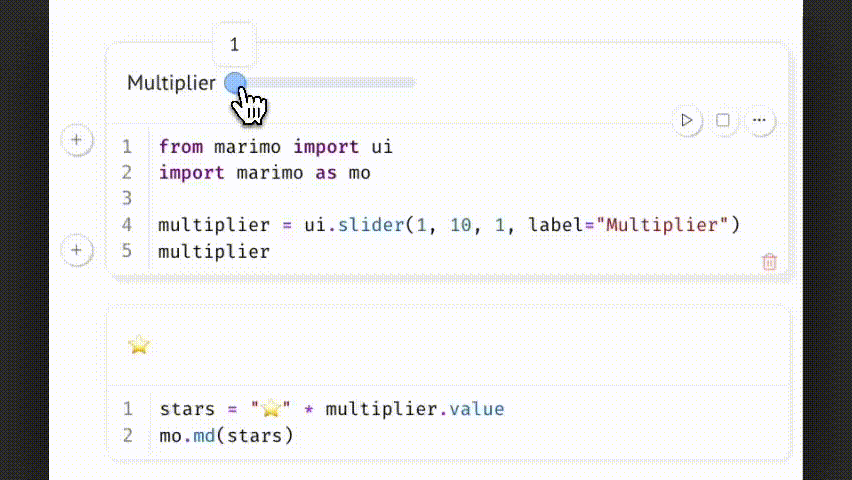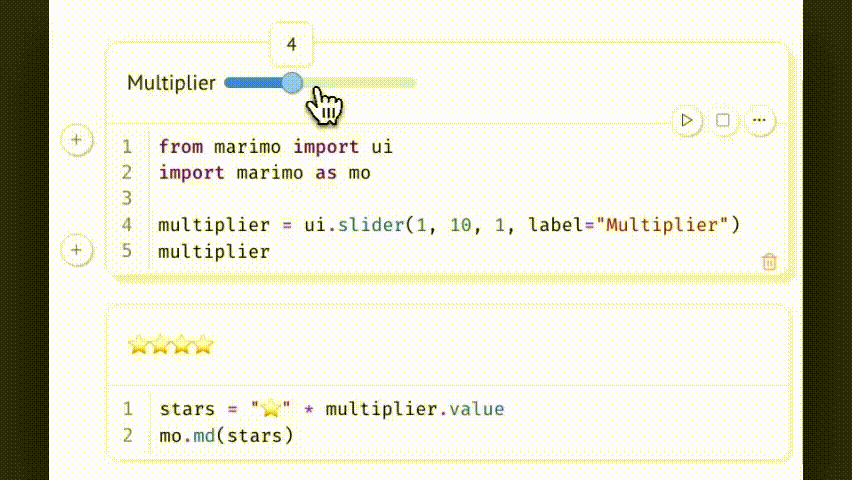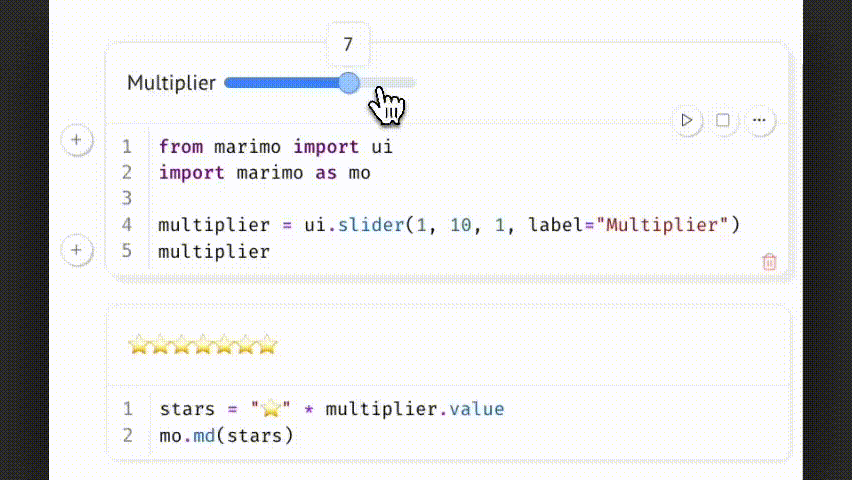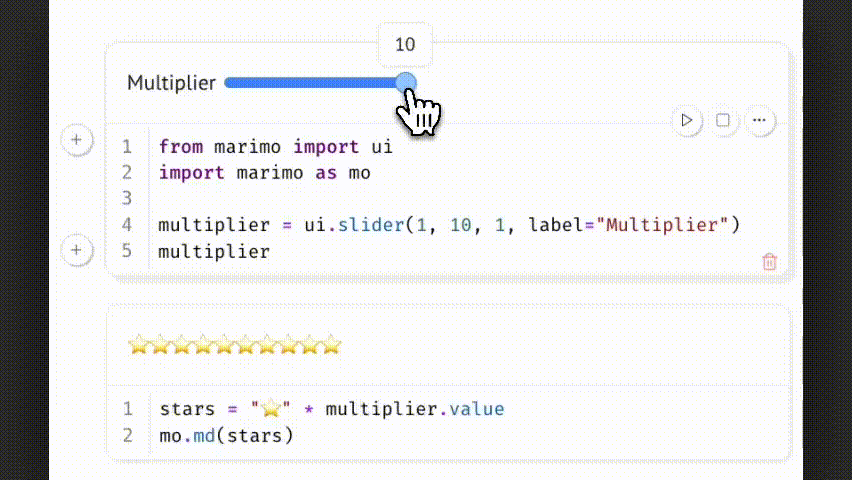
Changing the slider updates multiplier.value, which triggers all dependent cells. This allows parameter-driven exploration while preserving a reproducible and traceable code path.
Isolate Execution with Sandbox Mode
Problem in Traditional Notebooks
Notebook environments are not isolated by default, meaning dependencies or variables from other projects or past sessions can interfere with your results. There is no guarantee that what is in memory or on disk reflects only what is in the notebook, making reproducibility fragile.
How Marimo Solves This
Marimo offers a --sandbox mode that runs your notebook in a fully isolated environment. This ensures your code doesn’t unintentionally rely on globally installed packages or cached variables from other projects. Everything it needs must be declared in the notebook itself.

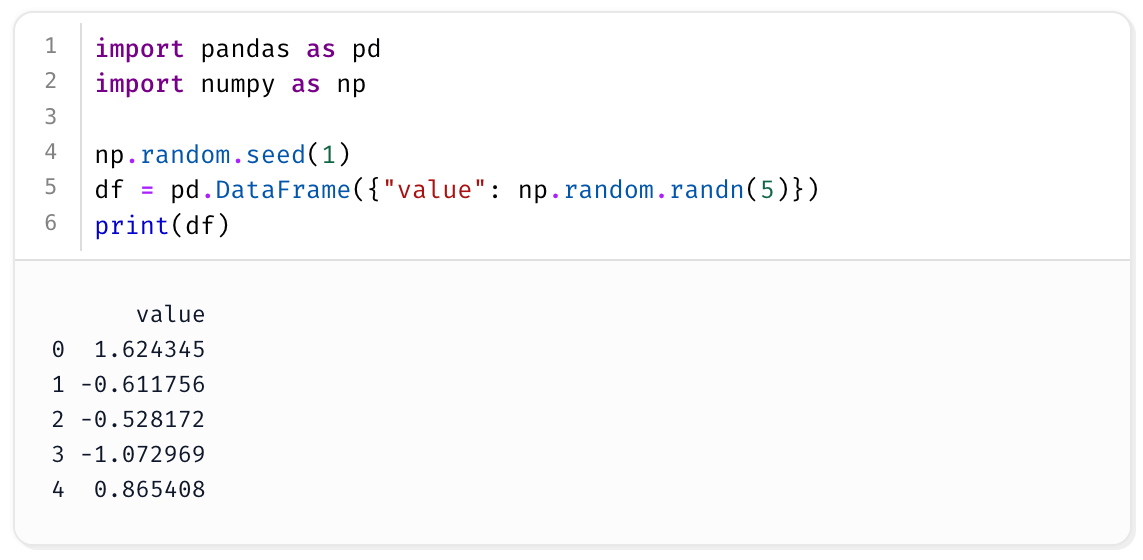
To see this in action, consider a minimal Marimo notebook:
import pandas as pd
import numpy as np
# Simulate some data
np.random.seed(1)
df = pd.DataFrame({"value": np.random.randn(5)})
print(df)
If pandas and numpy happen to be installed globally, you may not notice any issues while editing or running the notebook normally.
marimo edit my_notebook.py
But when you launch the notebook in sandbox mode, Marimo verifies that all dependencies are declared.
To use --sandbox, Marimo requires the uv package manager to manage isolated environments.
To edit the notebook in sandbox mode, run:
marimo edit my_notebook.py --sandbox
If any are missing, Marimo will prompt you to install them:
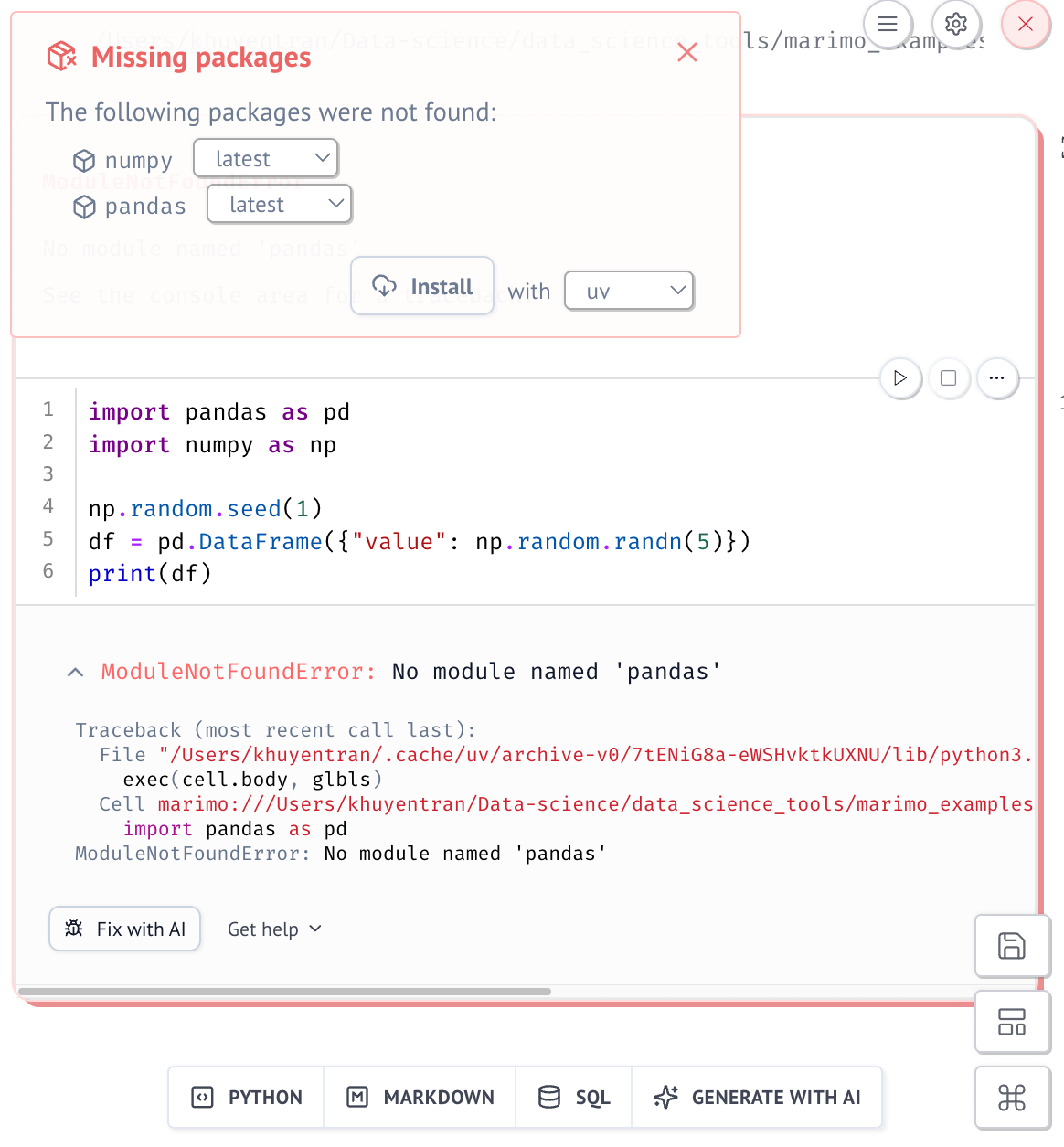
And then automatically add them to the notebook’s script header:
# /// script
# requires-python = ">=3.11"
# dependencies = [
# "marimo",
# "numpy==2.2.5",
# "pandas==2.2.3",
# ]
# ///
import marimo
__generated_with = "0.13.0"
app = marimo.App(width="medium")
@app.cell
def _():
import pandas as pd
import numpy as np
np.random.seed(1)
df = pd.DataFrame({"value": np.random.randn(5)})
print(df)
return
if __name__ == "__main__":
app.run()
The next time you open the notebook in sandbox mode, Marimo will reinstall these packages in an isolated environment before launching.
This ensures your notebook runs consistently everywhere, with no surprises. It’s a lightweight but powerful step toward full reproducibility.
Add Lightweight Unit Testing
Problem in Traditional Notebooks
In Jupyter, testing code requires manual effort or external scripts. There’s no built-in mechanism to validate the correctness of functions or outputs within the notebook itself, and you typically need to use workarounds like the ipytest package to integrate unit tests cleanly.
For example, using ipytest, you might write:
# Cell 1
import ipytest
import pytest
ipytest.autoconfig()
# Cell 2
def add(a, b):
return a + b
# Cell 3
%%ipytest -qq
def test_add():
assert add(2, 3) == 5
assert add(0, 0) == 0
While this allows inline testing, it introduces complexity and external tooling that Jupyter does not natively support.
How Marimo Solves This
Marimo automatically detects and runs test cells in your notebook. When pytest is installed, it runs tests from any cell that contains only test functions or classes that start with test_ or Test.
Example:
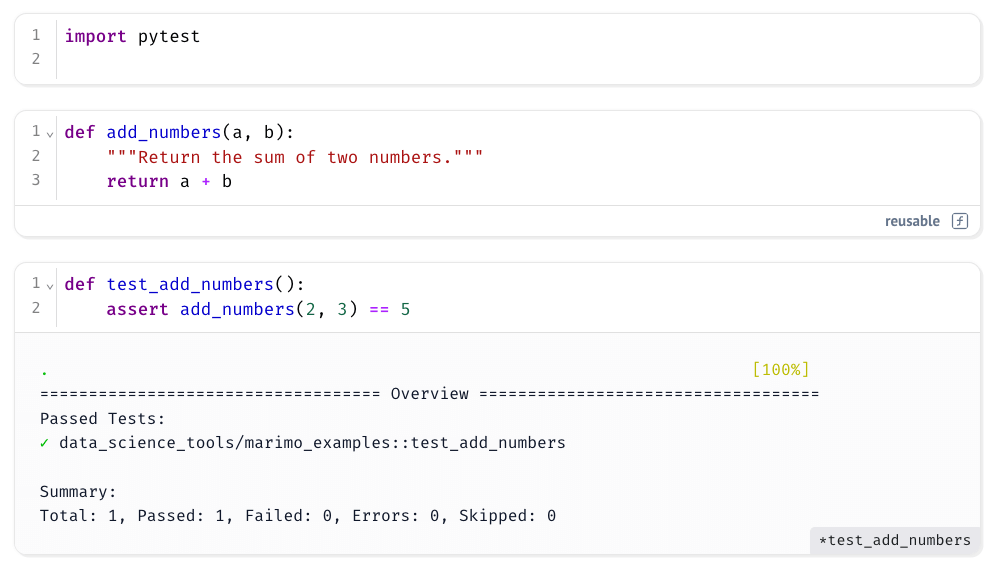
Running the test cell gives a summary of which tests passed or failed, along with their file and function names, just like a normal pytest session. This makes it easy to verify correctness within your notebook, without needing to leave your development environment.
You can also write parameterized tests with pytest.mark.parametrize for more coverage:
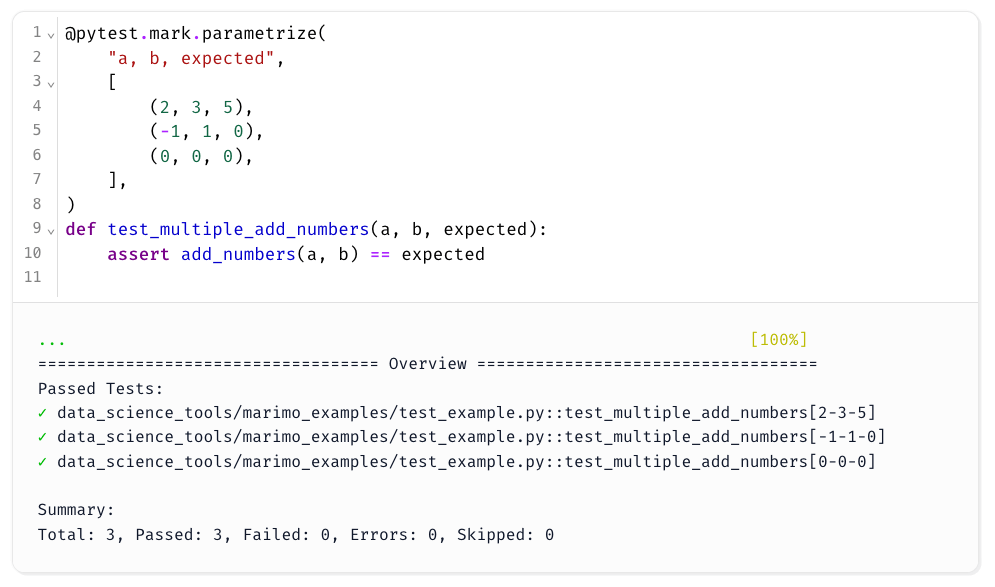
What’s even cooler is that because a Marimo notebook is just a Python script under the hood, you can run pytest directly on the notebook file from the terminal:
pytest test_example.py
Output:
test_example.py .... [100%]
=============================== 4 passed in 0.22s ===============================
This means you can test your notebook in a CI pipeline effortlessly, using standard pytest commands without any extra setup.
For a deeper dive into unit testing best practices tailored for data scientists, check out this article on pytest for data scientists, a great companion resource for building robust, testable notebooks.
Export in Multiple Reusable Formats
Problem in Traditional Notebooks
Notebooks are often treated as single-use or exploratory, making it difficult to modularize or reuse code without copying and pasting between files.
How Marimo Solves This
Marimo notebooks are not only readable, they’re also modular and exportable. You can convert your notebook into multiple formats depending on your use case in a data science project:
marimo export html my_notebook.py
marimo export html-wasm my_notebook.py
marimo export ipynb my_notebook.py
marimo export md my_notebook.py
marimo export script my_notebook.py # Clean Python script for production
Each export mode has a specific purpose:
- HTML: Great for sharing static visualizations and reports with stakeholders.
- HTML-WASM: Perfect for publishing interactive dashboards online without needing a backend server.
- IPYNB: Enables compatibility with traditional Jupyter workflows.
- Markdown: Ideal for documentation, blogs, or version-controlled notebooks in prose form.
- Script: Useful for integrating analytical logic into larger Python projects or production pipelines.
This flexibility allows you to deliver your analysis in the correct format for each stage of the data science lifecycle: exploration, communication, automation, or deployment.
Example Workflow: From Local Editing to Online Sharing
Let’s put everything together and explore how to go from local editing to an online shareable dashboard hosted on GitHub Pages.
You can find the source code for this example here, and the final dashboard, hosted on a GitHub page, here.
Edit Your Notebook in a Clean Environment
First, edit the notebook in sandbox mode:
marimo edit dashboard.py --sandbox
This ensures that all dependencies are declared, allowing the notebook to run independently of any system-level Python packages.
Export Interactive Results as a Shareable Dashboard
Next, export the interactive notebooks as a dashboard:
marimo export html-wasm dashboard.py -o build --sandbox
To launch a local web server for the folder, run:
python -m http.server --directory build
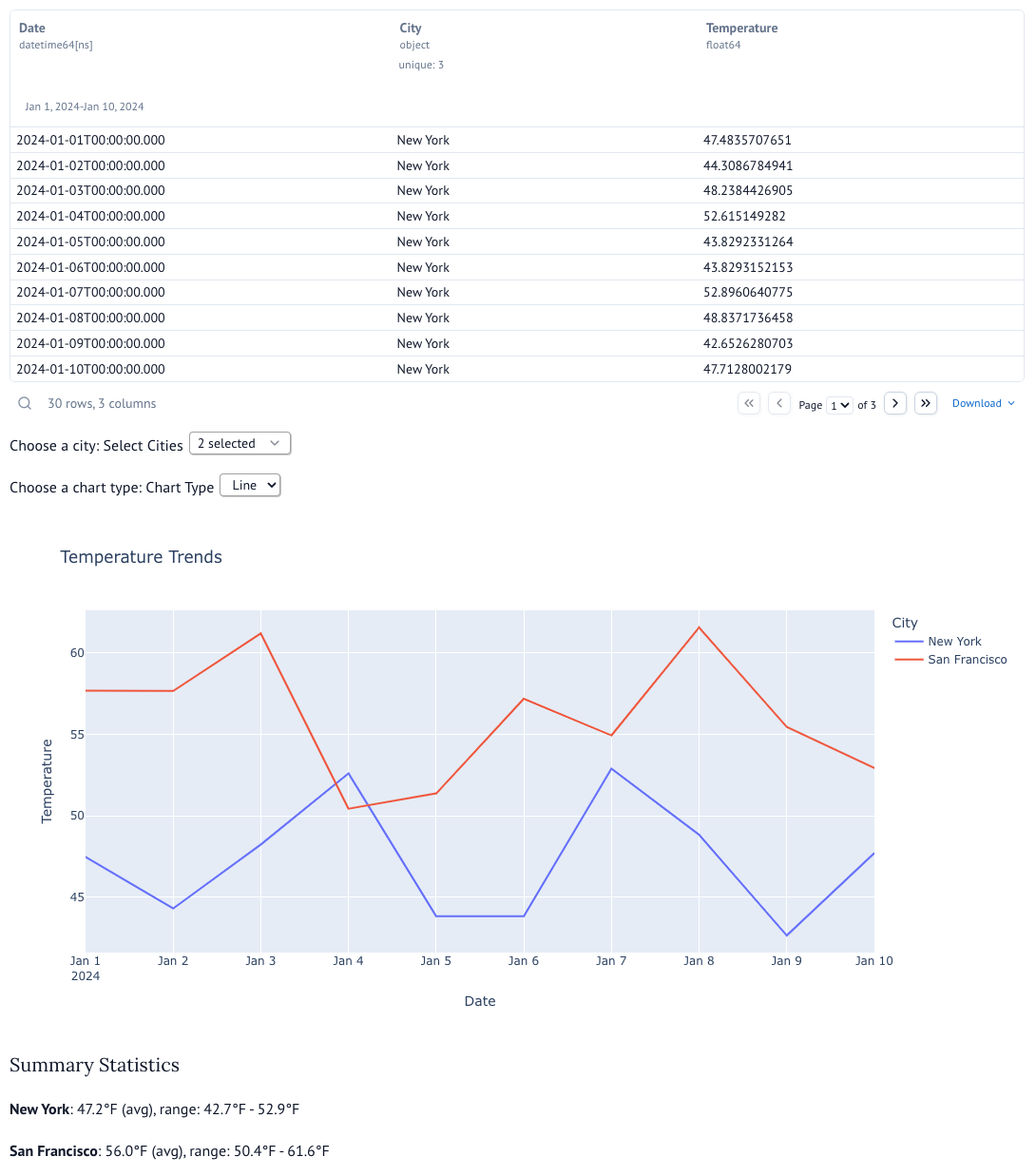
You will see an interactive dashboard as shown below:

Add a GitHub Actions Workflow
Let’s create a GitHub Actions workflow to rerun and export your notebook on every push:
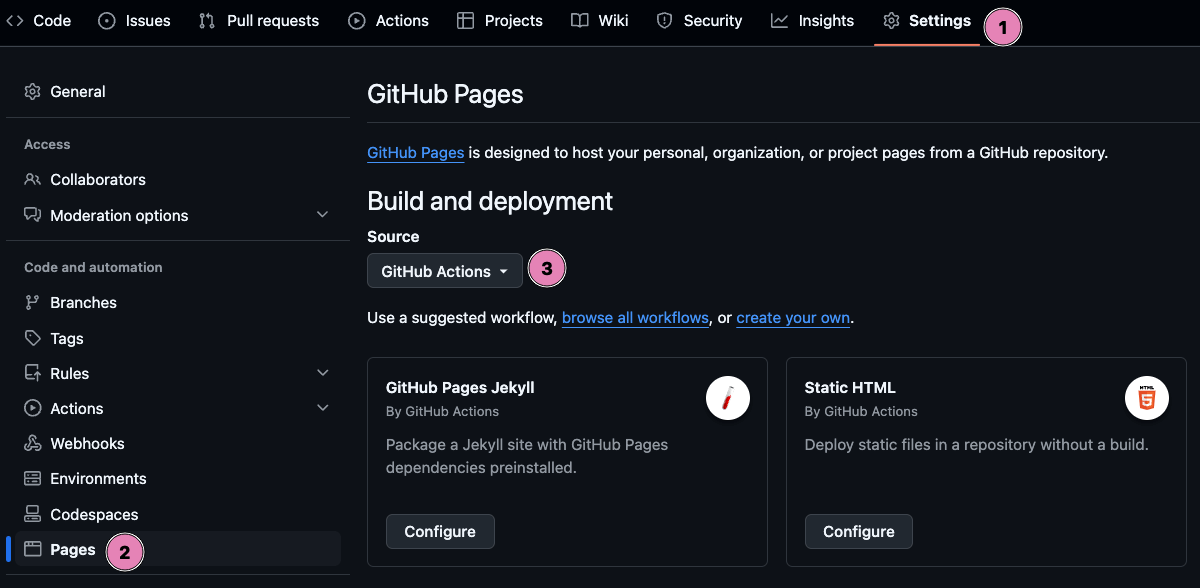
Start by configuring GitHub Pages for GitHub Actions by following these steps:
- Go to your repository and click the Settings tab.
- In the left sidebar, scroll down and click on Pages.
- Under Build and deployment, set the Source to GitHub Actions.
Add a GitHub Actions workflow file:
name: Deploy Marimo Notebook
on:
push:
branches: [main]
paths:
- dashboard.py
- .github/workflows/deploy.yml
jobs:
build:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v4
- name: Set up Python
uses: actions/setup-python@v4
with:
python-version: '3.11'
- name: Install uv
uses: astral-sh/setup-uv@v5
- name: Install dependencies
run: uv add marimo
- name: Export marimo notebook
run: uv run marimo export html-wasm dashboard.py --output build --sandbox
- name: Upload artifact
uses: actions/upload-pages-artifact@v3
with:
path: build
deploy:
needs: build
runs-on: ubuntu-latest
environment:
name: github-pages
url: ${{ steps.deployment.outputs.page_url }}
permissions:
pages: write
id-token: write
steps:
- name: Deploy to GitHub Pages
id: deployment
uses: actions/deploy-pages@v4
with:
artifact_name: github-pages
Add, commit, and push this workflow:
git add .
git commit -m 'add github workflow'
git push origin main
You should see something like this when visiting the website hosted on GitHub Pages.
Now, when you edit your Marimo notebook and push the changes to the GitHub repository, the GitHub page will automatically update with the modified dashboard.
Final Thoughts
Marimo brings reproducibility, shareability, and structure to data science notebooks.
If you’re tired of broken Jupyter workflows or want cleaner collaboration with Git, it’s worth exploring.
The combination of automatic dependency tracking, variable protection, clean version control, and multiple export formats makes Marimo a compelling alternative for data scientists who value both interactivity and reproducibility.
Related Resources
For deeper exploration of notebook development and reproducible workflows:
- Development Tools: Claude Code techniques guide for AI-assisted coding workflows
- Package Management: UV package manager guide for reproducible Python environments
- Logging: Loguru logging guide for data science project debugging
📚 Want to go deeper? Learning new techniques is the easy part. Knowing how to structure, test, and deploy them is what separates side projects from real work. My book shows you how to build data science projects that actually make it to production. Get the book →
Stay Current with CodeCut
Actionable Python tips, curated for busy data pros. Skim in under 2 minutes, three times a week.