Motivation
Data scientists and analysts often struggle with messy Git diffs when collaborating on Jupyter notebooks, as even small changes can result in large, hard-to-read differences due to cell outputs and metadata changes.
Example:
# Trying to understand changes in a .ipynb file diff
{
"cells": [
{
"cell_type": "code",
"execution_count": 1,
"metadata": {},
"outputs": [
{"data": {"text/plain": ["<large output blob>"]}, "execution_count": 1}
],
"source": ["import pandas as pd\n", "df = pd.read_csv('data.csv')"]
}
],
"metadata": {"kernelspec": {...}, "language_info": {...}}
}
Introduction to Jupytext
Jupytext is a tool that converts Jupyter notebooks to various text formats like Markdown documents or Python scripts, making it easier to control and edit versions in IDEs. It can be installed using pip:
pip install jupytextor conda:
conda install jupytext -c conda-forge
Paired Notebooks Feature
Jupytext solves the version control challenge by allowing you to pair your .ipynb files with text-based formats (.py or .md), maintaining both versions in sync automatically.
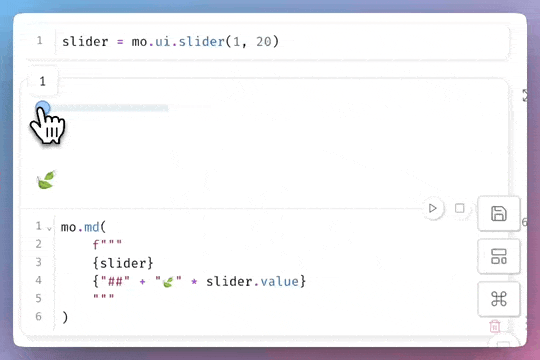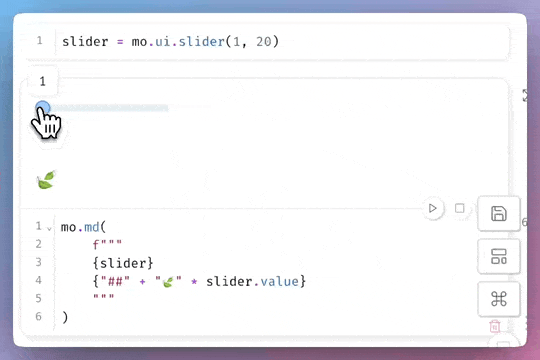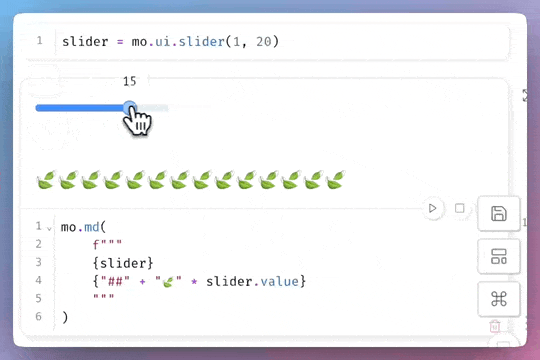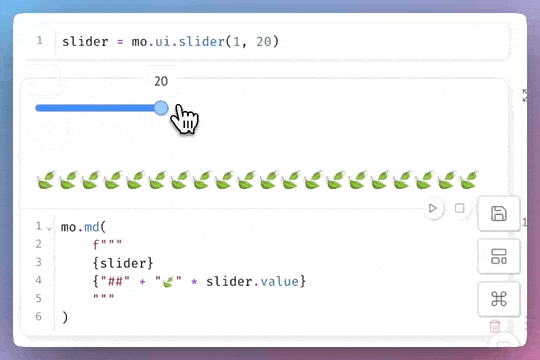
Let’s demonstrate how Jupytext automatically synchronizes paired files:
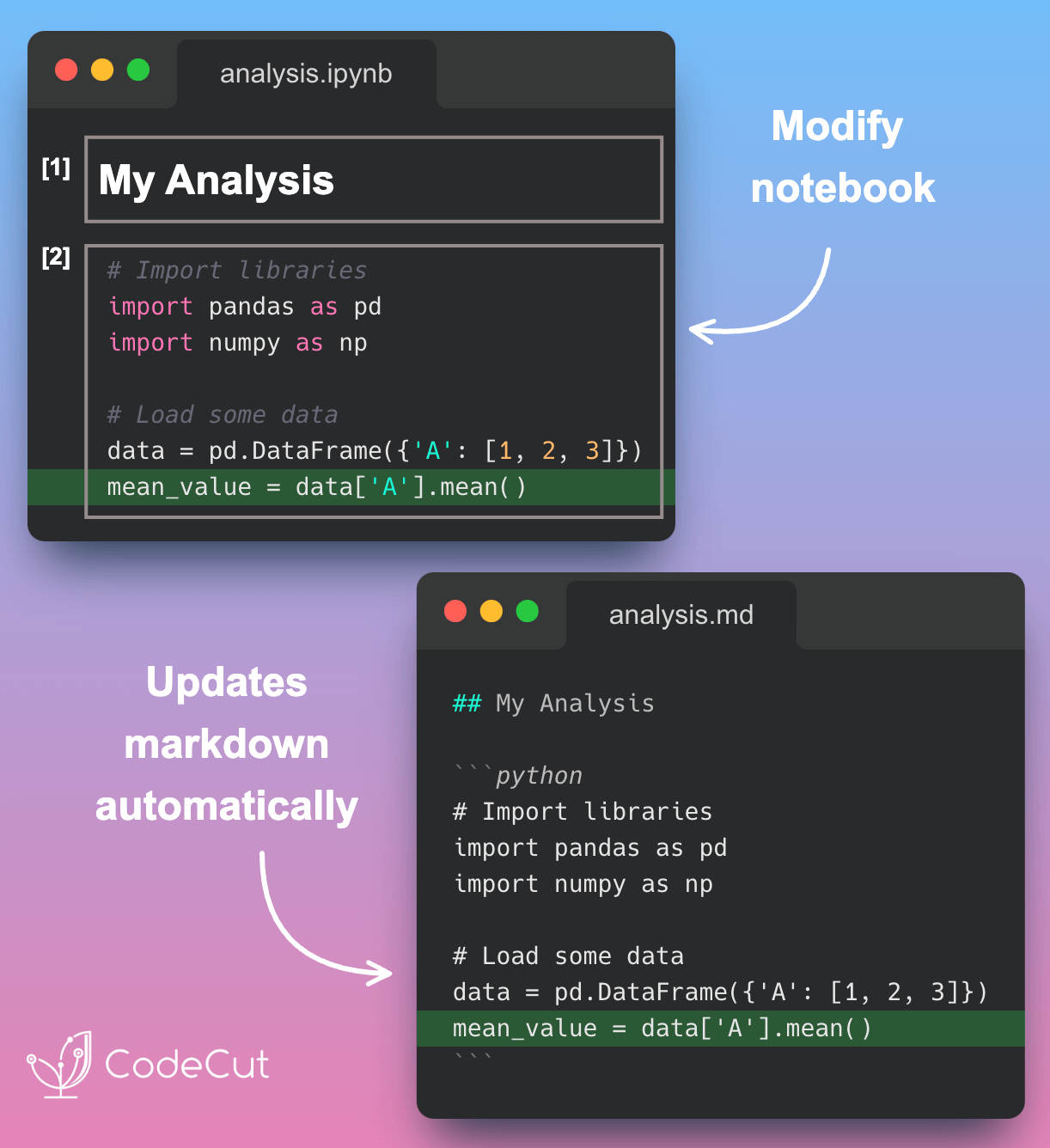
First, set up a paired notebook called analysis.ipynb:

In JupyterLab, pair your notebook to one or more text formats with the Jupytext commands:

This will automatically create a markdown version of the notebook called analysis.md:
---
jupyter:
jupytext:
formats: ipynb,md
text_representation:
extension: .md
format_name: markdown
format_version: '1.3'
jupytext_version: 1.16.7
kernelspec:
display_name: Python 3 (ipykernel)
language: python
name: python3
---
## My Analysis
```python
# Import libraries
import pandas as pd
import numpy as np
# Load some data
data = pd.DataFrame({'A': [1, 2, 3]})
```Now, let’s modify the notebook in Jupyter (analysis.ipynb):

After saving the notebook in Jupyter, analysis.md is automatically updated:
---
jupyter:
jupytext:
formats: ipynb,md
text_representation:
extension: .md
format_name: markdown
format_version: '1.3'
jupytext_version: 1.16.7
kernelspec:
display_name: Python 3 (ipykernel)
language: python
name: python3
---
## My Analysis
```python
# Import libraries
import pandas as pd
import numpy as np
# Load some data
data = pd.DataFrame({'A': [1, 2, 3]})
```
```python
# Add a new analysis section
mean_value = data['A'].mean()
print(f"Mean value: {mean_value}")
```You can also edit the markdown file directly and see changes reflected in the notebook:
---
jupyter:
jupytext:
formats: ipynb,md
text_representation:
extension: .md
format_name: markdown
format_version: '1.3'
jupytext_version: 1.16.7
kernelspec:
display_name: Python 3 (ipykernel)
language: python
name: python3
---
## My Analysis
```python
# Import libraries
import pandas as pd
import numpy as np
# Load some data
data = pd.DataFrame({'A': [1, 2, 3]})
```
```python
# Add a new analysis section
mean_value = data['A'].mean()
print(f"Mean value: {mean_value}")
```
## New SectionAfter reloading the notebook in Jupyter, analysis.ipynb will contain:

Key points about the synchronization:
- Changes in either file format are automatically reflected in the paired file when saving
- The synchronization preserves the notebook structure and cell types (code/markdown)
- Cell outputs are only stored in the
.ipynbfile - You need to reload the notebook in Jupyter to see changes made to the text file
This automatic synchronization makes it easy to:
- Edit notebooks in your preferred text editor
- Track changes in version control
- Collaborate with team members
- Maintain documentation and code in sync
Conclusion
Jupytext provides an elegant solution to version controlling Jupyter notebooks by separating the content from the outputs, making collaboration and code review much more manageable for data science teams. Its flexibility in supporting both Python and Markdown formats allows teams to choose the most appropriate format for their specific needs.